Molecular biological analysis of the genes coding the immunoglobulin (Ig) chains and the genes coding the antigen receptors of T-cells (T-cell receptor, TCR) are very useful methods which assist in the diagnostics and clasification of lymphoproliferative diseases.
In the years 1997-2008, we examined B or T -cells clonality in more than 8 000 patients.
B cells clonality
To understand the principle of the molecular genetic methods detecting the B-cells clonality it is firstly necessary to explain the mechanism of rearrangement of immunogobulin genes.
Rearrangement of the gene complexes encoding immunoglobulins.
Genetic information for immunoglobulins (Ig) is not inherited in a definitive form but in the form of gene segments. By joining these segments according to the development program of the organism during the lymphocyte differentiation, the appropriate complex genes are created. Their information is translated into the primary structure of one molecule of a light or heavy chain. Complex genes are set in the functional form only in B-lymphocytes where the rearrangement of the genes for heavy (IgH) and light (IgL) chains takes place. The process of their rearrangement is similar.
For the rearrangement, there is a general rule of allelic exclusion which, in the case of the gene for IgH, means that the process takes place only on one chromosome 14. We rarely find a biallelic rearrangement in tumor population. However, it have been described also the rearrangement in three alleles in the patient with a trisomy of chromosome 14.
Isotypic exclusion means that synthesis of the chains type κ and λ is mutually exclusive in each of the B-lymphocyte clones, i.e. there is the creation of only one type of light chain.
Rearrangement of the gene complex encoding the heavy chain of immunoglobuline (IgH).
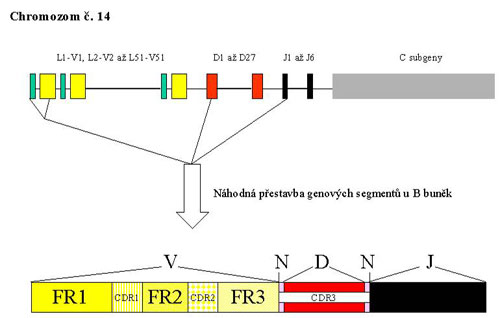
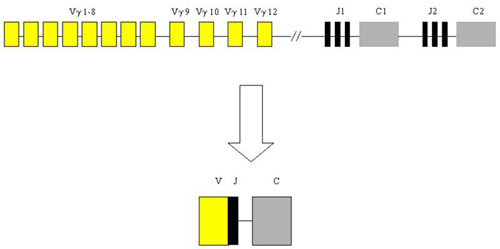
The gene complex for IgH is located on the long arm of chromosome 14 (in the region 14q32.33). The complex is composed of several types of segments which correspond to the appropriate parts of the heavy chain of the antibody molecule and so they carry the same designation - V - variable, D - ensuring diversity, J - joining and than C – constant. During the rearrangement of the IgH gene, there is randomly chosen one segment of each group from the multiple-member groups of segment which are then joined together. The rearrangement is a complicated process with a large possibility of combinations which is also supported by the inaccuracies in the joining of individual segments, resulting in a unique sequence of DNA. Somatic mutations in the immune response contribute to the diversity of B - lymphocytes and antibodies in the next stage of the development program.
Part of the gene complex coding the variable area of the antibody contains 4 conservative (framework) regions called FR I - FR IV. Among these, there are inserted 3 highly variable regions which assure the complementarity (complementarity determining regions, CDR). The rearrangement of IgH begins in an early pro-B-cell, when interuption in several parts of the gene complex takes place. Furthermore, there is always one segment chosen from the group 27 D and 6 J and they are joined together. The rearranged DJ section is then enlarged in a later pro-B-cell by adding one of the V segments (Fig. 1).
Figure 1
DNA sites of recognition are determined during the rearrangement by the sequences of 7 and 9 base pairs (heptamers and nonamers) which are recognized by two proteins activating recombination (recombination activating proteins 1, 2). Among nonamers and heptamers there are spacer sequences of 12 or 23 bp which correspond to one or two threads of DNA helix. On both ends of each segment, that should be involved in the rearrangement, there is one spacer sequence situated. Joining segments is only possible by the manner of the connection of 12 bp and 23 bp sequences. On both ends of the D segment there are 12 bp spacer sequences while in the 3´- end of the region V and in the 5´- end of the segment J there are 23 bp sequences. It follows that only the connection between D and J and V and D is possible, while V and J cannot be joined.
The variability and specificity of the rearrangement product is even more increased by the addition of nucleotides at both ends of the segment D (in the N zones) by terminal deoxynucleotidyl transferase. The resulting rearrangement is unique and characteristic for a concrete cell and for elements driven from it. To generate a complete code for the heavy chain of immunoglobulin a constant region has to be involved.
Adjunctive nucleotide insertions and deletions are not only the welcome source of variability. They can create a STOP codon or cause a frame shift so that the rearrangement is unproductive and does not allow the production of a functional protein. In that case, the process of rearrangement can be repeated on the second chromosome and if it is unsuccessful again the cell is damaged by apoptosis.
Rearrangement of the gene complexes encoding the light chain of immunoglobuline (IgL).
Gene complex encoding the kappa light chain is located on the chromosome 2 in the region p12 and information for the chain lambda is on the chromosome 22 in the region q11. Rearrangement of IgL takes place in the stage of pre-B-cells in a similar manner as the rearrangement of IgH. However, it is simpler because no D segments are present. After IgH rearrangement, the kappa rearrangement takes place first and the complex encoding lambda chain afterwards.
Importance of the examination of immunoglobulin gene rearrangement in diagnostics
The normal situation in the organism corresponds to polyclonality which means that there is a high amount of clones with different rearrangements. This allows the body to respond to a large amount of antigens. B-cell lymphomas represent the tumor proliferations outgoing from one transformed cell. Descendants of this cell have the same genetic make-up which can be identified in the area of the IgH by the Southern blot (after the cleavage with the appropriate restriction endonucleases) or by the means of a polymerase chain reaction (PCR). The first technique requires a great amount of high-molecular DNA and so its use in routine diagnosis is significantly limited. By contrast, thanks to the multiple amplification of a targeted DNA, PCR allows also the examination of archival material. Primers for PCR are complementary to the conservative regions of the IgH gene. These basic molecular-genetic examinations of lymphoproliferations help pathologists to resolve the question of malignancy and the question of the affiliation of pathologic process to B-cell or T-cell line.
Information about the monoclonality of pathological process helps in diagnosis of difficult cases, sometimes in the situation when other criteria (morphological and immunohistochemical) are difficult to evaluate as well, e.g. in expansive necrotic tissues. The identification of the monoclonal population significantly contributes to the diagnosis assessment of a malignant lymphoproliferation. However, it is still necessary to think about the fact that clonality is not always identical to malignancy. Monoclonal stripes may be also present when DNA is amplified from the tissue containing only a small number of lymphoid (albeit inflamatory) cells.
A population with a monoclonal rearrangement of IgH belongs mostly to B-cell line. In extensive studies of the immunoglobuline receptor and T-lymphocyte receptor there was also detected a phenomenon called the phenomenon of infidelity to an appropriate line ("lineage infidelity"). It happens when, for example, a tumor cell which according to other attributes belongs to the T-cell line shows a monoclonal IgH rearrangement.
Except for described applications in the examination of problematic biopsies, this sensitive examination of the IgH rearrangement by PCR is also used for the staging and monitoring of patients with B-cell lymphoma or leukemia and especially for monitoring the minimal residual disease.
Roughly speaking, the monoclonal process is mostly (but not always!) malignant and polyclonal process rather benign. In pathologic diagnosis it is of course always needed to evaluate the results of molecular genetics in context to other examinations.
The detection rate of method - what percentage of the real clonal lymphoproliferations is this method able to reveal? Generally it is between 60 % to 100 %. This, however, significantly depends also on the characteristics of the analyzed tissue. The high detection rate is in B-chronic lymphocytic leukemia / lymphoma of small B-lymphocytes, mantle-cell lymphoma and hairy-cell leukemia (up to 100 %) and much lower in follicular lymphoma, MALT lymphomas and acute lymphoblastic leukemia. Detection of IgH monoclonality using PCR is increased by the use of several types of reactions.
The sensitivity of method: population with a clonal IgH rearrangement should be revealed if forms at least 5 % of the sample cells.
Examination
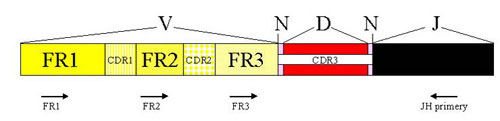
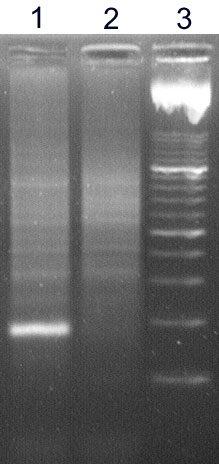
The analysis of IgH clonality in our laboratory is based on a polymerase chain reaction (PCR). PCR methods amplify the hypervariable segment of the VDJ region and use primers which are specific to the relatively conservative regions FR1, FR2, FR3 and JH (Fig. 2). The amplified products are then devided on agarose gel with a high resolution (high resolution agarose). The monolonal population creates a clear well-marked band, whilst the polyclonal population appears as a smear composed of fragments of various sizes (Fig.3)
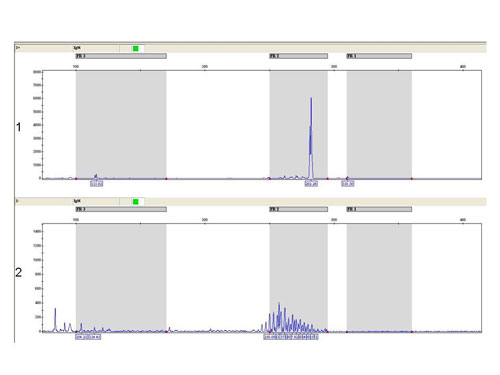
In debatable cases we use also the fragmentation analysis of fluorescently labeled PCR products (Fig. 4).
Figure 2
Figure 3
High resolution agarose gel: sample 1 - monolonal population creates a clear well-marked band, sample 2 - polyclonal population appears as a smear composed of fragments of various sizes, 3 - marker
Figure 4
The fragmentation analysis - FR2 reaction: sample 1: monoclonal population - one clear product, sample 2 - polyclonal population - more products
T – cells clonatity
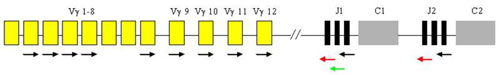
Lymphomas from T-cells are rare and represent only about 12 % of non-Hodgkin lymphomas. Diagnosis is difficult and requires the compilation of morphological, immunohistochemical and molecular-genetic methods. From the latter, the most important is the detection of T-cell clonality which utilizes the variability of T-cell antigen receptors (TCR). In principle, it is similar to the IgH method. The gene complex encoding TCR gamma in humans is localized on chromosome 7 in the region p15 - p14 and is composed of segments Vg, Jg and Cg which are randomly rearranged during T-cell development.
Examination
Analysis of T-cell clonality uses PCR amplification of the hypervariable region of the gene for the receptor of T-lymphocyte gamma (TCR gamma). We perform two different multiplex PCR with a mixture of fluorescently labeled primers which are complemetary to V and J segments. The fragmentation analysis of PCR products is performed on the genetic analyzer ABI PRISM 3130xl and evaluated by the use of GeneMapper (Fig. 7).
Figure 7
The fragmentation analysis - TCR gamma-B reaction: sample 1: monoclonal population - one clear product, sample 2 - polyclonal population - more products.
References
- Trainor KJ, Brisco MJ, Wan JH, Neoh S, Grist S, Morley AA. Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood. 1991;78(1):192-6.
- Aubin J, Davi F, Nguyen-Salomon F, Leboeuf D, Debert C, Taher M, Valensi F, Canioni D, Brousse N, Varet B, Flandrin and Macintyre EA. Description of a novel FR1 IgH PCR strategy and its comparison with three other strategies for the detection of clonality in B cell malignancies. Leukemia. 1995;9(3):471-9.
- Menke MA, Tiemann M, Vogelsang D, Boie C, Parwaresch R. Temperature gradient gel electrophoresis for analysis of a polymerase chain reaction-based diagnostic clonality assay in the early stages of cutaneous T-cell lymphomas. Electrophoresis. 1995;16(5):733-8.
- Miettinen M, Lasota J. Polymerase chain reaction based gene rearrangement studies in the diagnosis of follicular lymphoma--performance in formaldehyde-fixed tissue and application in clinical problem cases. Pathol Res Pract. 1997;193(1):9-19.
- Müller-Hermelink HK, Greiner A: Molecular analysis of human immunoglobulin heavy chain genes (IgVH) in normal and malignant B cells. Am J Pathol. 1998;153(5):1341-6.
- World Health Organisation Classification of Tumours: Pathology and Genetics. Tumours of Haematopoietic and Lymphoid Tissues. Ed. Jaffe, ES, Harris, NL, Stein, H, Vardiman JW. IARC Press, Lyon, 2001.
- Ahnhudt C, Muche JM, Dijkstal K, Sterry W, Lukowsky A. An approach to the sensitivity of temperature-gradient gel electrophoresis in the detection of clonally expanded T-cells in cutaneous T-cell lymphoma. Electrophoresis. 2001;22(1):33-8